Liquid biopsy to diagnose esophageal adenocarcinoma
This impressive multicenter study by Miyoshi et al. introduces a new diagnostic tool named EMERALD, which is a six-circulating miRNA signature designed to effectively differentiate patients with esophageal adenocarcinoma (EAC) and its precursor, Barrett's esophagus (BE), from non-disease controls. The research involved analyzing 792 patient samples across four countries to develop and validate the blood-based assay. RNA was isolated from tissue and serum samples, and miRNA expression was analyzed using quantitative PCR. Candidate biomarkers were selected based on specific statistical criteria from The Cancer Genome Atlas (TCGA) dataset. A total of nine miRNAs were identified as significantly over-expressed in EAC tissues compared to non-diseased controls. The study then transitioned to a blood-based assay, confirming that six of these miRNAs could be detected in serum and were upregulated in EAC patients. The diagnostic potential of these miRNAs was assessed using machine learning models (XGBoost and AdaBoost) and a Markov model was employed to simulate screening strategies for patients with chronic gastroesophageal reflux disease (GERD). The findings suggest that these miRNAs could serve as non-invasive biomarkers for early detection of EAC and its precursors. Furthermore, a cost-effectiveness analysis suggested that implementing this liquid biopsy-based screening approach at five-year intervals could facilitate earlier diagnoses and reduce mortality rates compared to traditional endoscopy methods.
Liquid biopsy to identify Barrett's oesophagus, dysplasia and oesophageal adenocarcinoma: the EMERALD multicentre study.
Miyoshi J, Mannucci A, Scarpa M, Gao F, Toden S, Whitsett T, Inge LJ, Bremner RM, Takayama T, Cheng Y, Bottiglieri T, Nagetaal ID, Shrubsole MJ, Zaidi AH, Wang X, Coleman HG, Anderson LA, Meltzer SJ, Goel A; FINBAR-EMERALD collaborative group.
Abstract
Background: There is no clinically relevant serological marker for the early detection of oesophageal adenocarcinoma (EAC) and its precursor lesion, Barrett's oesophagus (BE).
Objective: To develop and test a blood-based assay for EAC and BE.
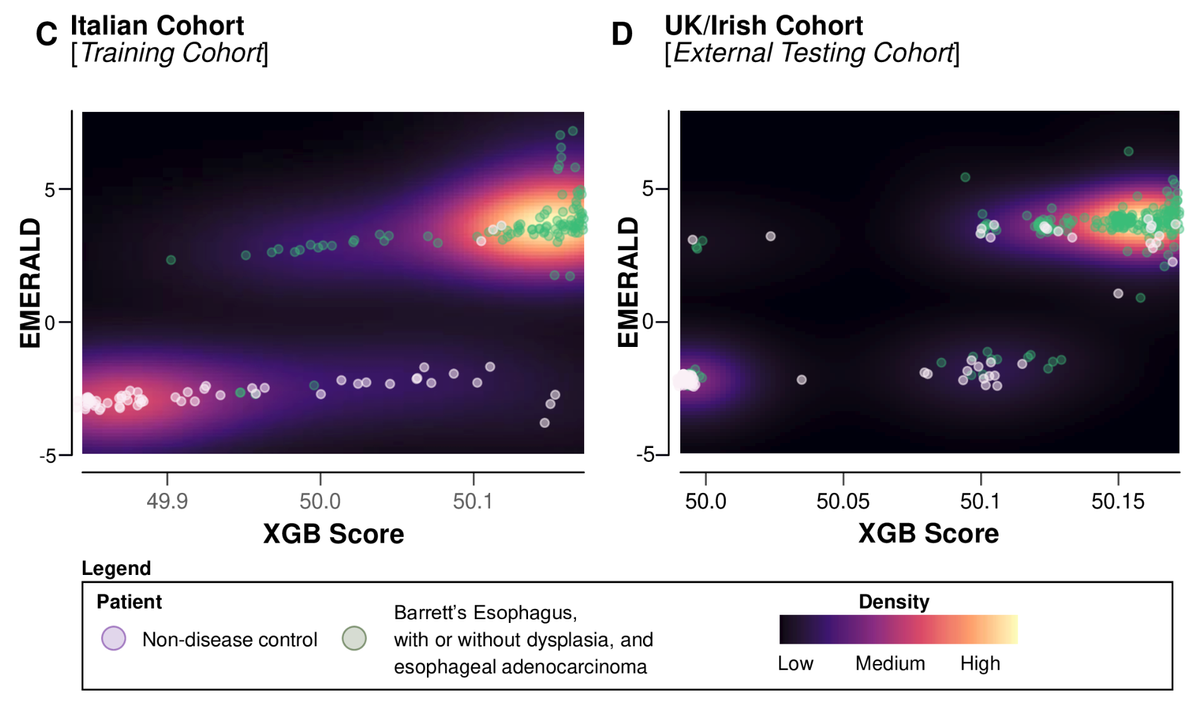
Design: Oesophageal MicroRNAs of BaRRett, Adenocarcinoma and Dysplasia (EMERALD) was a large, international, multicentre biomarker cohort study involving 792 patient samples from 4 countries (NCT06381583) to develop and validate a circulating miRNA signature for the early detection of EAC and high-risk BE. Tissue-based miRNA sequencing and microarray datasets (n=134) were used to identify candidate miRNAs of diagnostic potential, followed by validation using 42 pairs of matched cancer and normal tissues. The usefulness of the candidate miRNAs was initially assessed using 108 sera (44 EAC, 34 EAC precursors and 30 non-disease controls). We finally trained a machine learning model (XGBoost+AdaBoost) on RT-qPCR results from circulating miRNAs from a training cohort (n=160) and independently tested it in an external cohort (n=295).
Results: After a strict process of biomarker discovery and selection, we identified six miRNAs that were overexpressed in all sera of patients compared with non-disease controls from three independent cohorts of different nationalities (miR-106b, miR-146a, miR-15a, miR-18a, miR-21 and miR-93). We established a six-miRNA diagnostic signature using the training cohort (area under the receiver operating characteristic curve (AUROC): 97.6%) and tested it in an independent cohort (AUROC: 91.9%). This assay could also identify patients with BE among patients with gastro-oesophageal reflux disease (AUROC: 94.8%, sensitivity: 92.8%, specificity: 85.1%).
Conclusion: Using a comprehensive approach integrating unbiased genome-wide biomarker discovery and several independent experimental validations, we have developed and validated a novel blood test that might complement screening options for BE/EAC.